Bacterial pathogens of plants and animals are related
Type III secretion system in bacterial plant and animal pathogens
Pathogens in different biological kingdoms rely on effector proteins to cause disease
Most microbes are not pathogens
Where do bacteria go inside the plant?
How does the plant recognize invading bacteria?
What happens after the plant surface receptors detect microbial features?
How is the microbial recognition event communicated from surface receptors so the plant can make all these defense responses?
What has been learned by deleting all the Type III effector genes and adding them back?
| Bacterial pathogens of plants and animals are related |
|
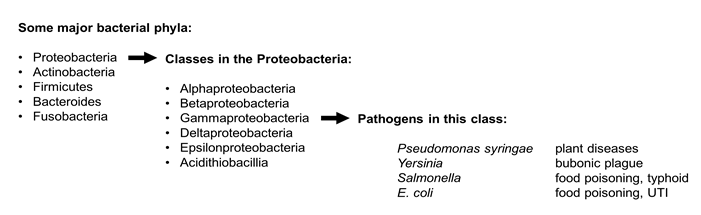
Bacteria are divided into different phylogenetic groups based on sequence comparisons of core genes such as those that encode ribosomal RNA. Many important bacterial pathogens are members of the phylum Proteobacteria. Proteobacteria are divided into several classes. Pseudomonas syringae, the plant pathogen that is the focus of the Vegevaders(TM) game is a members of the class called Gammaproteobacteria. Gammaproteobacteria also include familiar animal pathogens like E. coli, Salmonella, and Yersinia.
| The Type III secretion system in bacterial plant and animal pathogens |
|
Bacterial pathogens of plants and animals share similar strategies for causing disease. Research conducted on one type of pathogen can help researchers better understand how other pathogens cause disease.
For example, bacterial pathogens of plants and animals both use the Type III secretion system to secrete effector proteins into their plant and animal hosts. The type III secretion system is a complex apparatus related to the bacterial flagella and was first described in animal pathogens like Salmonella and Yersinia. This information was very useful to plant pathologists as they began to characterize the Type III secretion system in Pseudomonas syringae and other plant pathogens.
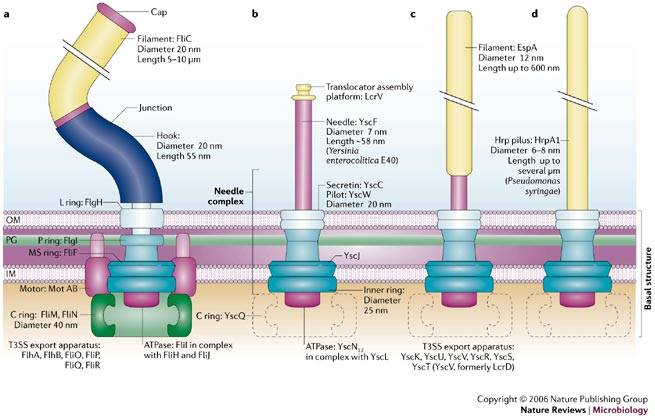
Now we know that type III secretion systems fall into different groups having slightly different overall structures. The figure below compares the structures of (a) a bacterial flagella with type III secretion systems from (b) Yersinia, (c) E. coli, and (d) Pseudomonas syringae.
Figure 1 below is taken from a review article:
Cornelis, G.R. 2006. The type III secretion injectisome. Nat Rev Micro 4:811-825.
You can read the whole article here: http://www.ncbi.nlm.nih.gov/pubmed/17041629
Figure 1.
| Pathogens in different biological kingdoms use effector proteins to cause disease |
|
Effector proteins are defined as proteins secreted from or exposed on the outside of the pathogen from where they interact with the host organism. As discussed in the previous section, many bacterial pathogens secrete effectors with the type III secretion system. But effectors are produced by many different pathogens and not just bacteria!
Living organisms are currently classified as belonging to one of six different kingdoms (http://en.wikipedia.org/wiki/Kingdom_(biology)). Plant pathogens are found in the kingdoms Bacteria, Fungi, Protista, and Animal. Pathogens in all four of these kingdoms use effector proteins to interfere with diverse plant cell functions including plant defense pathways.
Table 1.
| Pathogen type |
Kingdom |
Effector entry into the host plant |
Read more about these specific effectors |
| bacteria |
Bacteria |
Effectors are injected through the plant cell wall into the cytoplasm |
HopU1
made by the Pseudomonas syringae (Fu 2007)
XopD
made by Xanthomonas campestris (Kim 2008) |
| fungi |
Fungi |
Effectors are secreted into the apoplast (plant intracellular space). Some of these are translocated across the membrane into the plant cytoplasm |
Pep1
made by the corn smut fungus Ustilago maydis (Hemetsberger 2012) |
| oomycetes |
Protista |
Effectors are secreted into the apoplast. Some of these are translocated across the membrane into the plant cytoplasm |
AVRblb2
made by the Irish potato famine pathogen Phytophthera infestans (Bozkurt 2011) |
| nematodes |
Animals |
Effectors are secreted into the apoplast or injected through the plant cell wall into the cytoplasm |
GrCEP12
made by the potato cyst nematode Globodera rostochiensis (Chen 2013) |
| Most microbes are not pathogens |
|
Plants and animals are surrounded by microbes, most of which don't cause disease at all. In fact, many of the microbes that live in and on our bodies are beneficial to our health. And there are lots of them! By some estimates our bodies have 10 times more microbial cells than human cells.
Plants also have microbes living in and around them. Some, like the bacterium Rhizobium, have been known for a long time and benefit their host plants by providing them with nitrogen.
Many of the microbes that live in association with plants and animals can't be easily grown on artificial media, so only recently, with the power of cheap and fast gene sequencing, are we beginning to get a picture of the true microbial diversity. In humans, characterization of the microbes in and around us has been conducted by the Human Microbiome Project. In this study, 15-18 sites on the bodies of 242 people were sampled. A profile of the bacteria present at each site was generated by sequencing the 16S ribosomal RNA genes. Sites that were sampled included the belly button, the inner elbow and the web between the toes. (http://www.nature.com/nature/focus/humanmicrobiota/)
Figure 2.
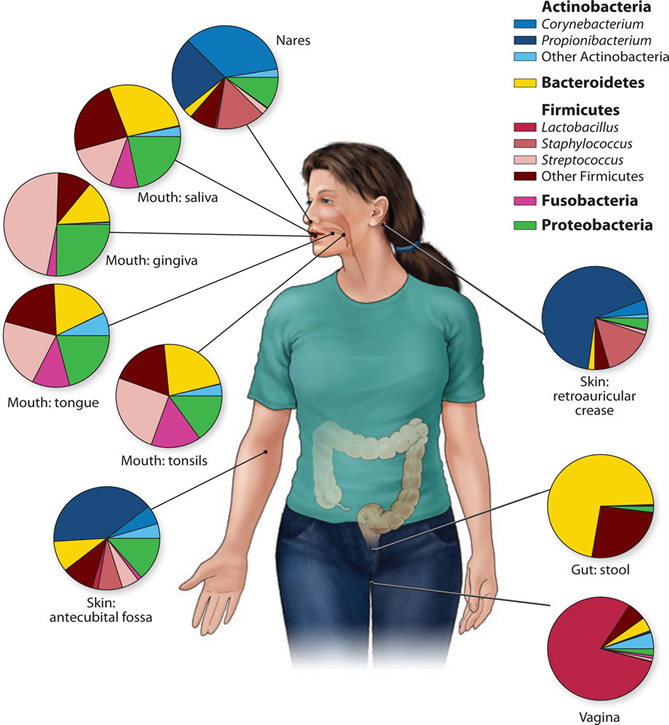
Genus- and phylum-level classification of bacteria colonizing a composite subject, showing that human microbiome diversity is dependent on the site sampled. Sites in the oral cavity share greater similarity than other types of sites, such as the skin, vagina, and gut.
Figure credit: Grice, E.A., and Segre, J.A. 2012. The human microbiome: our second genome. Annual review of genomics and human genetics 13:151-170. Read more: http://www.ncbi.nlm.nih.gov/pubmed/22703178
Research on plant microbiomes is less advanced, but experimental approaches similar to those being used with humans is being used to characterize microbes living in and around plant roots and inside the plant itself. It is hoped that microbes can be identified that provide nutritional and defensive benefits to the plant. Manipulation of plant-associated microbes may represent an important tool for increasing agricultural yields.
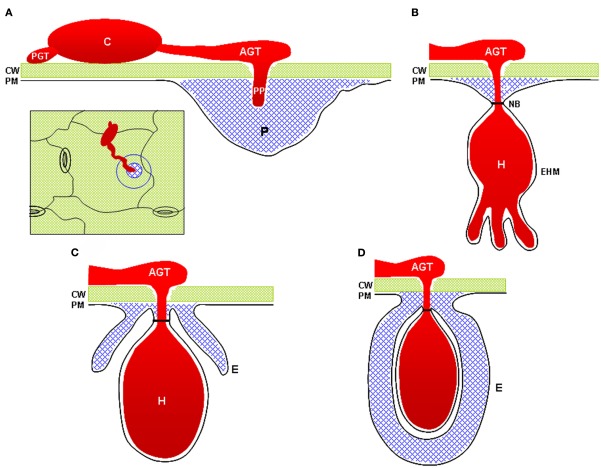
| Bacteria inside the plant |
|
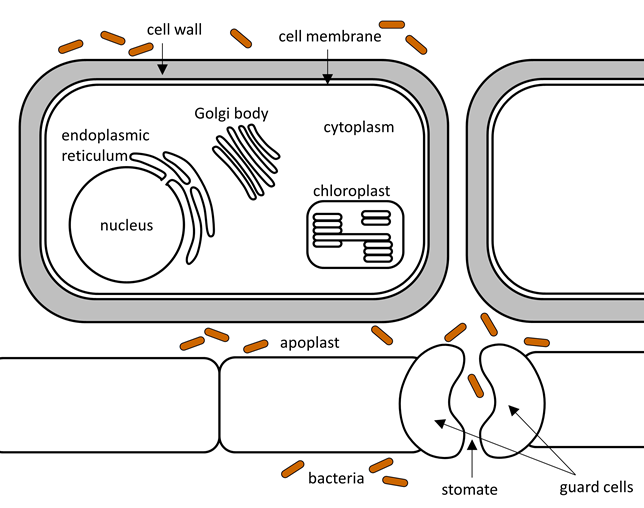
Bacteria enter the plant through wounds or stomates. They are then free to move through the apoplast that surrounds the plant cells, but they can't cross the thick plant cell walls.
Figure 3.
| How does the plant recognize invading bacteria? |
|
The plant identifies the invading bacteria by using receptor proteins that detect conserved microbial features. This strategy works very well because the conserved features are typically important to the survival of the bacteria. Figure 4 shows locations of some of the recognized features. Table 2 lists microbial features, the names of their plant receptor proteins, and links to scientific papers on these interactions.
Figure 4.
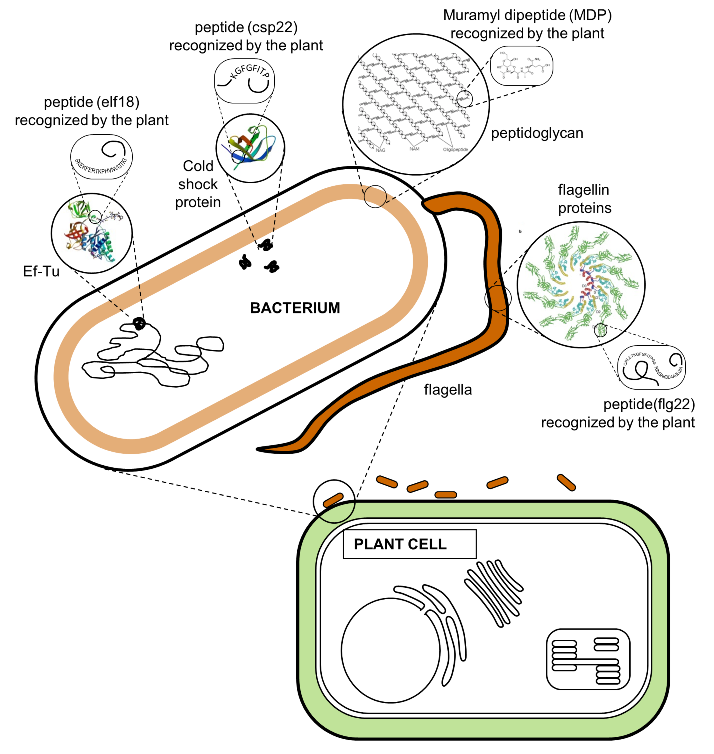
Table 2.
| Microbial Feature |
Role in the bacteria |
Plant Receptor |
Interesting detail |
Read more |
| flagellin |
Required for movement |
FLS2 |
flagellin perception is especially important in tomato plants |
Chinchilla, 2006 |
| Elongation factor Ef-Tu |
required for translation of mRNA into protein |
EFR |
Ef-Tu perception is especially important in the mustard family (Brassicaceae) |
Lacombe, 2010 |
| peptidoglycan |
part of the bacterial cell wall |
Lym1 and Lym2 |
both plants and higher animals can detect bacterial peptidoglycan
|
Willmann, 2011 |
| cold shock protein |
Regulates transcription and translation |
unknown |
|
Felix, 2003 |
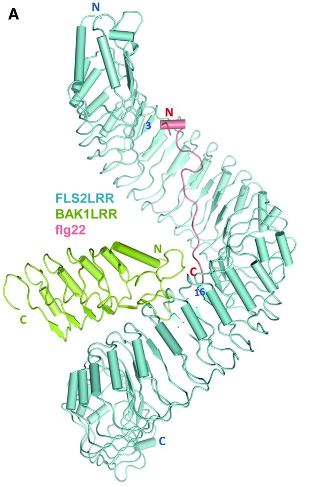
Plant receptors bind microbial features directly by a highly specific "lock-and-key" interaction. One of the best characterized of the plant receptors is FLS2. FLS2 is a receptor-like kinase protein located in the cell membrane just inside of the plant cell wall. When FLS2 binds to the flg22 peptide in flagellin (Fig 4), it triggers association with another protein called BAK1 and initiation of signalling pathways in the plant cell that lead to defense.
In 2013, the structure of the FLS2-BAK1-flg22 protein complex was solved using X-ray crystallography (Sun et al, 2013. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342:624). A diagram of this structure is shown in Figure 5. You can read the whole paper at: http://www.ncbi.nlm.nih.gov/pubmed/24114786.
Figure 5. Plant surface receptor (FLS2) binds bacterial feature (flg22)
| What happens after the plant surface receptors detect microbial features? |
|
How do plants defend themselves? They can't run away or growl and bare their teeth. They don't make antibodies like we do. Kind of like a medieval castle defending itself, plant defense involves closing the locations where pathogens can enter (stomates), strengthening the walls (callose), and making defensive "weaponry" that is damaging to the pathogen (such as phenolic compounds).
Stomatal closure:
As diagrammed in Fig 3 (above), stomates are one of the main entry points for pathogens.
Figure 6 shows an open stomate on a tomato leaf (left) and a tomato leaf stomate that has closed after detection of the bacterial pathogen Pseudomonas syringae pv. tabaci (right).
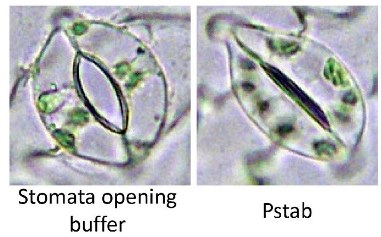 . .
This figure is taken from the paper by Lee et al (Coronatine inhibits stomatal closure and delays hypersensitive cell death induced by nonhost bacterial pathgens. PeerJ 1:e34.) http://www.ncbi.nlm.nih.gov/pubmed/23638370
Strengthening the cell wall:
the plant cell wall reporesents an important preformed barrier to pathogen invasion. Many plants have been shown to strengthen or reinforce cell walls upon detection of bacterial features These reinforced areas are referred to as cell wall appositions of papillae and one of their main components is the polysaccharide callose. Callose is frequently used as an indicator of whether a microbe or microbial feature is inducing immunity
Figure 7. Microscopic visualization of stained callose deposits in Arabidopsis thaliana. The photo on the left shows a high level of callose induced in response to bacterial features. When the plant has a mutation that interferes with signaling after the recognition of microbial features (as in the NahG plant on the right) much less callose is produced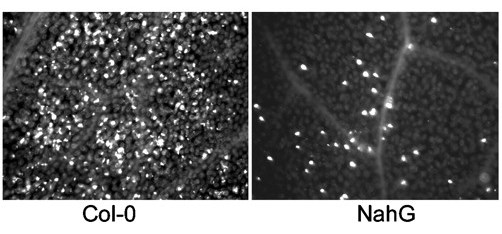
This figure is taken from a paper by Debroy et al 2004 (A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plant. PNAS 101:9927)
http://www.ncbi.nlm.nih.gov/pubmed/15210989
Figure 8. Cartoon showing how strengthening the plant cell wall can interfere with pathogen entry. Some fungi (red) are able to directly penetrate the plant cell wall (green). Callose-containing cell wall apositions (gray) surround the fungal structures and halt further development
This figure is taken from a review by Underwood 2012 (The plant cell wall: a dynamic barrier against pathogen invasion. Frontiers in Plant Science 7:85) http://www.ncbi.nlm.nih.gov/pubmed/22639669
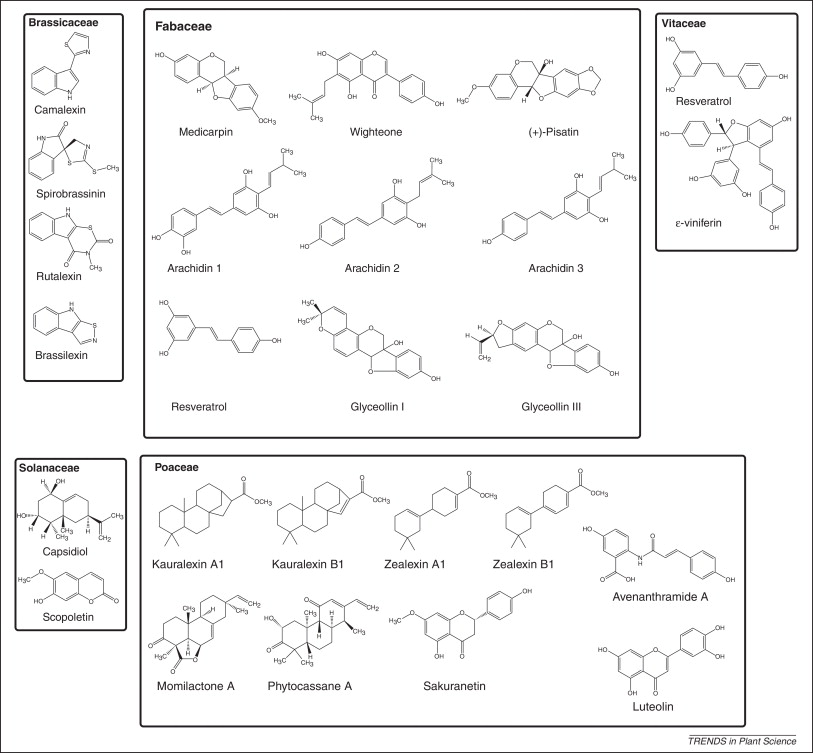
Defense compounds:
Detection of microbial features also results in the production of defense compounds includin antimicrobial non-protein toxins called phytoalexins. Figure 9 shows the structures of some phytoalexins produced by different plant families ( Brassicaceae = mustard family, Fabaceae = beans, Poaceae = grains, Solanaceae = potatoes/tomatoes/peppers, Vitaceae = grapes). Notice that one of the grape phytoalexins is Resveratrol for which there is some evidence for positive effects on human health.
Figure 9 is taken from a review article by Ahuja et al 2012 (Phytoalexins in defense against pathogens, Trends in Plant Sci 17:73). http://www.ncbi.nlm.nih.gov/pubmed/22209038.
| How is the microbial recognition event communicated from surface receptors so the plant can make all these defense responses? |
|
Detection events are communicated from upstream receptors to downstream targets by MAP kinase cascades. Kinases are enzymes which add a phosphate group to another protein. MAP kinases are serine/threonine /tyrosine kinases. MAP kinase cascades are composed of three types of kinases - the MAPKKK, MAPKK, and MAPK which are activated in response to external stimuli and lead to activation and suppression of transcription factors so that the right set of genes in turned on to respond to the stimulus (read more here).
MAP kinase cascades were first characterized in animal systems, but plants produce many more. Two of the best studied MAP kinase cascades in plants have been characterized in Arabidopsis and are shown in Figure 10.
Several P. syringae type III effectors suppress the plant defense response by disrupting the normal function of MAP kinase cascades. Details of how they do this are shown in Table 3.
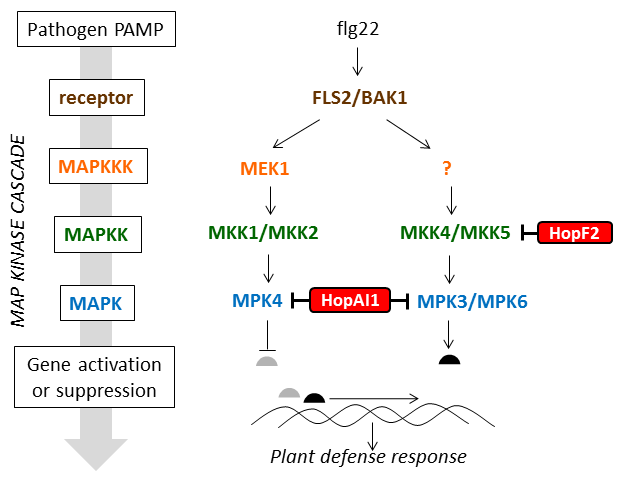
Figure 10 is adapted from a review article by Pitzschke, 2009 (MAPK cascade signalling networks in plant defence, Current opinion in Plant Biology 12:421) http://www.ncbi.nlm.nih.gov/pubmed/19608449
Table 3
| Type III effector name |
Enzymatic activity |
Impact on MAP kinases |
Read more |
| HopAI1 |
phosphothreonine lyase |
removes the phosphate group from the threonine residue required for activation of MPK3, MPK4, and MPK6 by their upstream MAPKKKs |
Zhang 2007, Zhang 2012 |
| HopF2 |
ADP-ribosyltransferase |
ADP ribosylates MKK5 |
Wang 2010 |
| What has been learned by deleting all the Type III effector genes and adding them back? |
|
Many plant pathogens, such as the bacterial speck pathogen Pseudomonas syringae pv. tomato DC3000, inject multiple effectors (nearly 30 in this case) into host cells. Removing and separately studying all of these components of the virulence system has revealed key effectors and their functions. Some effectors (HopG1, HopE1, HopM1, HopAM1) disrupt basic cellular functions to interfere with plant defenses, promote an aqueous environment in the air spaces within a leaf (where bacteria grow), and produce disease symptoms. Effector AvrPtoB, in contrast, targets the plant’s defensive recognition systems and is like a Swiss Army knife in how it does this. AvrPtoB disrupts both pattern-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI involves surface recognition of universal microbial features like flagellin. ETI is aimed at pathogen effectors inside plant cells and can be triggered by one of AvrPtoB’s own protein domains or by other effectors in the same repertoire. AvrPtoB can block self-domain ETI by using its “E3” activity to destroy a plant protein needed to recognize that domain. However, E3 mutants become better at suppressing the ETI triggered by other effectors in the same repertoire, thus revealing two competing ETI suppressing activities in one effector. This knowledge is helping us to develop tomato plants with better resistance to pathogens. Figure 11 is modified from a review article by Wei and Collmer 2017 (Defining essential processes in plant pathogenesis with Pseudomonas syringae pv. tomato DC3000 disarmed polymutants and a subset of key type III effectors. Mol. Plant Pathol. 19, 1779) https://www.ncbi.nlm.nih.gov/pubmed/29277959

|






 .
.



